The following section outlines the activities in which UC3M and FIBHGM participate.
APR Selection
We developed a deep learning approach to automatically estimate the patietnt position from a photograph captured prior to X-ray exposure, enabling the automatic selection of optimal prime factors (kVp, mAs, and source-detector distance) that directly influence radiation dose and image quality.
To build and evaluate the model:
- A database of 66 common radiographic positions was created from 75 volunteers across two X-ray facilities.
- A lightweight ConvNeXt architecture was trained with fine-tuning, discriminative learning rates, and a one-cycle scheduler.
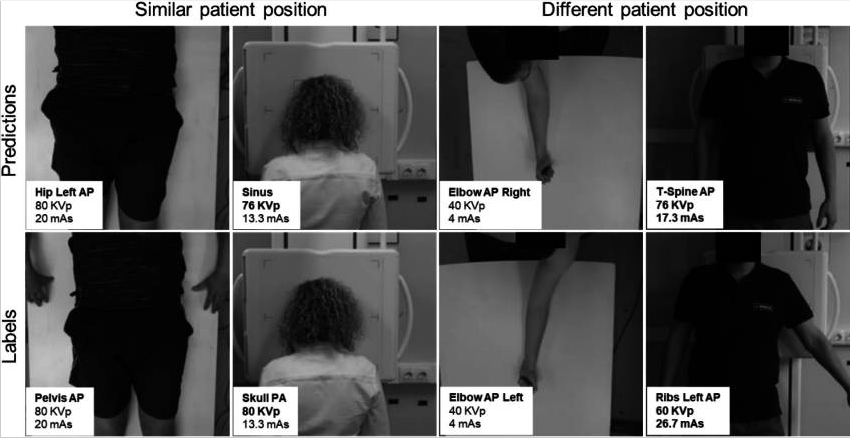
The proposed model achieved an accuracy of 93.17% for radiographic position classification, which increased to 95.58% when evaluating the correct selection of prime factors, as half of the errors corresponded to positions sharing the same kVp and mAs values. Most misclassifications occurred in positions with similar patient poses, suggesting that visual similarity remains a challenging factor. These results demonstrate the feasibility of the method to streamline the acquisition workflow, reduce exposure errors, and minimize unnecessary radiation dose.
The figure below illustrates representative scenarios with similar poses (left) and different poses (right). Errors are highlighted in bold.
These results were published in the Journal of Imaging Informatics in Medicine in 2024, under the title: Deep Learning–Based Estimation of Radiographic Position to Automatically Set Up the X-Ray Prime Factors
Automatic Collimation
During the acquisition of a radiological image, the X-ray beam is collimated to limit the irradiated region and thus the ionizing radiation delivered to the patient. This collimation, which depends on both the radiological position to be acquired and the patient morphology, is set manually by a technician before the acquisition. An incorrect collimation may result in a degradation of the image contrast, additional radiation dose to the patient or an insufficient field of view.
A Deep Learning-based method has been developed to automatically perform the collimation of the X-ray beam. The optimal aperture of the collimator is automatically inferred by analyzing an image of the patient —either a photograph, a 3D scan, or a combination of both— taken right before the acquisition. This differs from most previous works in the literature, where the collimation is defined on low-dose radiographs, requiring extra radiation dose.

The few non-ionizing methods in the literature are tailored to work on just anteroposterior abdominal and leg studies, while we extend our analysis to of the most common radiological positions. Moreover, we employ state of the art Neural Architecture Search methods to generate very tailored architectures that set a high bar for what can be accomplished following the proposed approach.

Our results are consistent with inter- and intra-operator variability, and show that the proposed method can be used in real systems to facilitate the acquisition workflow, reducing exposition. The preliminary results from this line of research have been published in the CASEIB 2023 under the title Colimación automática para sistemas de radiología mediante aprendizaje profundo, and the latest advancements are currently being condensed into a journal paper.
Automatic Quality Control
Three problems related to the quality of X-ray images have been addressed: excessive noise, inadequate contrast, and patient positioning errors.
To study the case of excessive noise, the degradation of radiographs has been simulated by adding quantum noise (dependent on amperage) and electronic noise (Gaussian), training a model that can identify images of insufficient quality.

Inadequate adjustment of the amperage (mAs) or the presence of non-ideal detector responses can modify the relationship between tissue attenuation and recorded gray levels, influencing the intensity distribution in the image. The expected contrast variability in radiographs was simulated using exponential histogram transformations.

Finally, in the incorrect patient positioning case, CT volumes were used, rotating them in a range of [-30º, 30º] with respect to an ideal orientation, and projecting the corresponding radiographs.

For all three cases, two similar approaches were followed: (1) creating a regression model for each variable of interest—μ for Poisson noise, Δγ for contrast transformation with respect to ideal γ, and degrees of rotation, respectively—setting a warning threshold when the predicted variable falls outside the acceptable range, and (2) directly training the models for multiclass classification, applying this threshold to the training data instead of the model output—with the classes [underexposed, acceptable] for the noise problem, [underexposed, acceptable, overexposed] for the contrast problem, and [rotated to the left, correct, rotated to the right] for the rotation problem. Both approaches have produced almost indistinguishable results, so the former has been chosen as the main approach as it provides more information than the binary class.
Image Enhancement
Denoising
We developed DDPM-X, a method for noise removal in planar radiographic images based on a diffusion model. This method involves two main stages. In the first stage, a diffusion model is trained with real clinical data to generate images by progressively removing Gaussian noise, starting from a fully noisy input. In the second stage, the method determines the most appropriate point in the generative chain to begin denoising real images. This is achieved by identifying the step that corresponds to the noise level present in the given image.
The proposed method was evaluated on both simulated Poisson (DDPM-Xg) and Gaussian noise (DDPM-Xp), at a high and a low dose, and on real noise.
Our results show that, unlike the other methods to which we compare it, the proposed model can denoise large radiographic images while preserving resolution, with the following advantages:
- Despite having been trained specifically for Gaussian noise, the diffusion model can effectively handle Poisson noise removal without requiring adjustments or retraining.
- Given the conditioning of the noise level in the network, the user can regulate the amount of noise removal in an ad hoc manner.
High dose

|
Model |
LPIPS ↓ |
SSIM ↑ |
PSNR ↑ |
|
BM3D |
0.02 |
98.88 |
39.78 |
|
Vanilla UNet |
0.02 |
98.68 |
41.39 |
|
Nei2Nei |
0.02 |
98.80 |
40.45 |
|
DU-GAN |
0.02 |
98.11 |
42.23 |
|
DDPM-Xg |
0.01 |
99.15 |
42.54 |
|
DDPM-Xp |
0.01 |
99.07 |
41.93 |
Low dose:

|
Model |
LPIPS ↓ |
SSIM ↑ |
PSNR ↑ |
|
BM3D |
0.05 |
97.81 |
34.58 |
|
Vanilla UNet |
0.07 |
97.81 |
37.56 |
|
Nei2Nei |
0.03 |
96.85 |
36.45 |
|
DU-GAN |
0.04 |
97.61 |
37.04 |
|
DDPM-Xg |
0.02 |
97.33 |
37.27 |
|
DDPM-Xp |
0.02 |
98.05 |
36.89 |
Real noise
The image below shows denoising results for an anthropomorphic phantom acquired at 100 kV / 4 mAs and 100 kV / 0.8 mAs. The red arrow indicates missing details.

These results were published in 2024 in the Proceedings of Machine Learning Research journal, under the title Diffusion X-ray image denoising
Contrast and Resolution Enhancement
For contrast and resolution improvement, we developed radBoost, an efficient algorithm implemented in the C programming language. To reduce execution time, a multithreaded implementation based on OpenMP was developed. The execution time of the conventional algorithm is 2 seconds for 4000×4000 pixel images, so acceleration on GPU-type devices has not been considered. The proposed algorithm uses the CLAHE method (Contrast Limited Adaptive Histogram Equalization) to enhance contrast and a 3-level Laplacian pyramid to improve resolution. To avoid enhancing undesired elements that may appear in the background, radBoost includes a preliminary segmentation step using the triangle algorithm. Due to the wide range of patient sizes in veterinary imaging, the method has been adapted to ensure robustness against varying image contrasts. This has been achieved through the development of different configurations that can be tailored depending on the clinical context and user preferences.
The image below shows radiography images prior to enhancement with radBoost (left) and after (right):

These resutls were published in CASEIB 2022 under the title Corrección automática del contraste en imagen radiográfica mediante aprendizaje profundo.
Reduction of Tissue Overlap
Bone Supression
We proposed a method based on Stable Diffusion, a text-to-image generative model pre-trained on natural images, which was first retrained using Textual Inversion to generate radiographs with bone suppression, and then conditioned with a ControlNet-type network to generate bone-free images from radiographs. As a baseline, we used a U-Net architecture, using ResNet34 as the encoder and comparing three loss functions: MSE, MSSIM, and LPIPS.
Results show that our approach achieves full bone suppression while preserving spatial detail, outperforming traditional Deep Learning methods in perceptual metrics. As a limitation, the appearance of minor hallucinations in soft tissue regions may limit clinical interpretation. While the method is currently limited to CXRs of 512×512 px, it can easily be expanded to larger sizes by using higher resolution versions of Stable Diffusion.
The image below shows, from left to right: CXR, U-Net, proposed method, and bone-suppressed CXR. The second row corresponds to a zoomed-in view of the region indicated by the yellow rectangle. Red arrows indicate hallucinations or loss of anatomical details.

| Metric | UNet | SD + ControlNet |
| MSSSIM ↑ | 94.32 | 91.96 |
| PSRN ↑ | 31.34 | 29.53 |
| LPIPS ↓ | 0.11 | 0.08 |
| CPBD ↑ | 0.37 | 0.48 |
These results were published in 2025 in Medical Imaging with Deep Learning-Short Papers under the title Bone supression in planar X-ray images with Stable Diffusion
Reconstruction
Conventional tomosynthesis reconstruction provides pseudo-tomographic information but still suffers from tissue superposition. To address this limitation, we proposed a filtered back-projection step (FDK) followed by deep learning-based post-processing to obtain a tomographic image closer to CT quality. To enhance the FDK reconstruction, four deep learning post-processing strategies were evaluated:
-
Single-step 2D post-processing: A modified U-Net was applied on individual planes: axial (2DAx), coronal (2DCor), and sagittal (2DSag).
-
2.5D post-processing: Combined the outputs of 2DAx, 2DCor, and 2DSag by averaging them to integrate information from all planes
-
Multi-step 2D post-processing: Applied two correction steps sequentially on orthogonal planes (e.g., Ax2Cor, Cor2Ax, Sag2Ax)
- 3D patch-based post-processing: Evaluated two 3D models: 3D U-Net (3D-P-U) that uses 3D convolutions for volumetric learning and 3D UneTR (3D-P-UTr): integrates attention mechanisms for better spatial representation.
Single-step 2D post-processing
Figure below shows the result of the 2D approaches. The blue arrow shows a high density mass in the lungs. The red arrow shows discontinuities in the
single-step 2D volumes. It can be observed that 2D approaches introduce discontinuities on planes not used during training and often miss lung structures, except 2DSag, which recovered more details.

Muilti-step 2D and 3D post-processing
While multi-step 2D post-processing reduces visible discontinuities compared to single-step methods, it cannot fully restore fine details lost in earlier stages. Among the tested variants, Sag2Ax achieved the best balance, managing to recover the lung mass, whereas Ax2Cor and Cor2Ax failed to do so.3D-P-U shows the best visualization of the overall shape and structure of the ground truth volume but fails to recover the high-density mass in the lungs. In contrast, 3D-P-UTr manages to preserve part of this mass while maintaining good structural continuity.

Overall, the 2D post-processing of sagittal slices showed high potential for reconstructing fine details within the lungs, while 3D patch-based post-processing achieves the best reconstruction of the overall patient shape. Preliminary results demonstrated the potential of the proposed methodology to obtain true tomographic information from tomosynthesis data. These results were presented in the 8th International Conference on Image Formation in X-ray Computed Tomography (“CT Meeting 2024”).
X-Ray Simulation
A modular software framework has been developed for X-ray simulation, offering flexibility to design and execute customizable simulation workflows from chest CT volumes. The framework supports three imaging modalities: planar radiography, digital tomosynthesis and CT.

The framework is organized into three main modules, corresponding to the following simulation stages: (1) preprocessing, where the input chest CT volume is transformed into a patient-specific model, (2) projection, where X-rays projections are generated based on acquisition geometry with optional physical effects and noise, and (3) reconstruction, where the volume is generated from the simulated projections. The figure below shows an example workflow of the simulation stages tailored for digital chest tomosynthesis.

The framework decouples projection operations from physical modeling, delegating projection and backprojection tasks to external engines, which can be easily integrated through the framework’s standardized interface.
Given the absence of complete end-to-end simulation frameworks for tomosynthesis, a version of this work, focused exclusively on tomosynthesis simulation and including a dedicated wrapper for the ASTRA Toolbox was presented in the 38th IEEE International Symposium on Computer-Based Medical Systems (CBMS) in 2025, under the title ChestXsim: An Open-Source Framework for Realistic Chest X-Ray Tomosynthesis Simulations. This version is planned for public release in an open repository, together with instructions and example workflows for a user-friendly execution. Additionally, a manifest with a curated set of downloadable CT scans from the Medical Imaging and Data Resource Center (MIDRC) database is provided to ensure reproducibility.
Overall, the developed simulation framework represents a valuable tool for generating paired imaging datasets that are otherwise difficult to obtain in clinical practice. It enables the creation of synthetic images under controlled conditions, suitable for training deep learning algorithms for noise reduction, artifact correction, or reconstruction enhancement.
Diagnostic Support
Automatic Nodule Detection
An automatic tool has been developed for the detection of pulmonary nodules from computed tomography (CT) images and simulated tomosynthesis projections. These are reconstructed using the FDK method, generating lower-resolution volumes, and subsequently enhanced by a Deep Learning model (DL-CT) that approximates their quality to that of a real CT. Detection is carried out with a U-Net network trained on CT slices, adapted to FDK and DL-CT through transfer learning, and cross-inference is also evaluated by directly applying the CT model on DL-CT data.
Evaluated on a test set, the model trained on CT remains the reference, although it shows some loss of generalization. Models with transfer learning exhibit limited performance, especially in detecting small nodules, while cross-inference with DL-CT provides a better balance:
- +5.7% in precision and –5.4% in sensitivity compared to CT
- +10% in small nodule detection compared to FDK
Figure below shows two representative examples of predictions obtained by the different models (in yellow) compared with the ground truth labels (in blue):
- Top row: large nodule clearly visible in CT and FDK, correctly detected by all models.
- Bottom row: nodule poorly visible in FDK. Models with transfer learning fail to detect it, while cross-inference with DL-CT partially recovers the detection, but with a smaller estimated size.

Overall, DL-CT is presented as a promising reconstruction method for tomosynthesis, capable of approaching the performance of conventional CT and improving small nodule detection compared to the FDK method, while maintaining tomosynthesis advantages such as lower radiation dose and easier acquisition for chest studies.
Distribution Matching
A distribution matching method has been proposed based on a combination of Deep Learning and classical computer vision, capable of transforming computed tomography data taken with different acquisition parameters (FOV, amperage, background objects, etc.) into a common space. This allows different databases to be combined to obtain large amounts of information and train deep learning models for any task of interest. In addition, it ensures the generalization of the models to any future input data distribution, transforming the latter into the common space as a form of preprocessing.

Likewise, an interface has been designed for the automatic segmentation of structures—specifically designed for lungs, but generalizable to any other problem—so that users without a high level of programming skills can access powerful deep learning tools. The tool allows users to make inferences and train or retrain models with just a couple of clicks. Along with the interface, pre-trained models are provided to apply the distribution matching method defined above (using YOLOv11), as well as to perform fibrotic lung segmentation (using a U-Net with ResNet50 as an encoder).
 The method and resulting user interface have been published in the CASEIB 2024 congress under the title Segmentación robusta y automática de pulmón fibrótico en modelo animal, and has been expanded since for its use in both preclinical and clinical applications.
The method and resulting user interface have been published in the CASEIB 2024 congress under the title Segmentación robusta y automática de pulmón fibrótico en modelo animal, and has been expanded since for its use in both preclinical and clinical applications.
Neural Architecture Search
Neural Architecture Search (NAS) is a field of Machine Learning that aims to automatize the model development workflow. Recent advancements in the field particularly focus on generating and deploying Deep Learning models, from the exploration of different architectures to the optimization of their hyperparameters. NAS is rapidly gaining traction, as it excels in
- multiobjective optimization, balancing the tradeof between smaller (for simpler, cheaper hardware), faster (for real time applications) and more accurate (for crucial medical problems) models,
- novel architecture generation, exploring the vast range of posibilities for Deep Learning model design and discovering new functional blocks that outperform the state of the art in scarce but complex data scenarios and
- low skill requirements for use, as automatizing the whole pipeline from model creation to deployment allows non-experts in Deep Learning (and even non-programmers) to access novel architectures tailored to their specific needs.
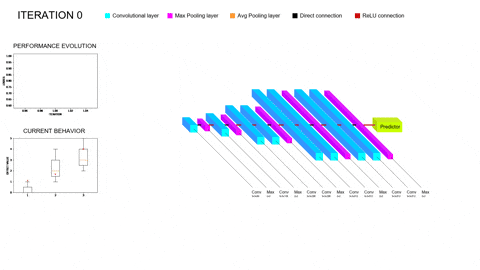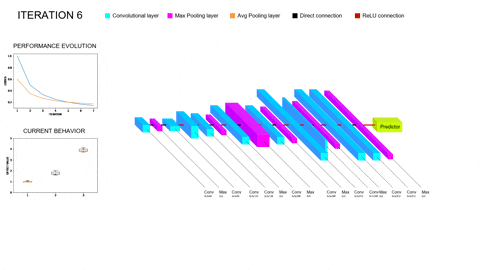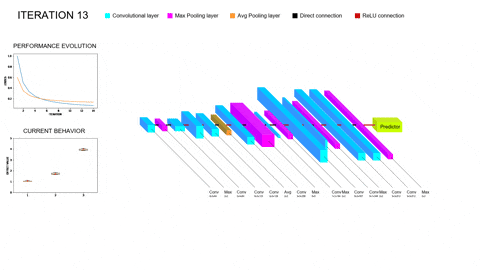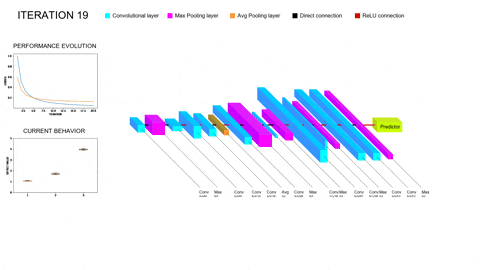
Although promissing, these methods bring their own set of problems to the table. On the one hand, training a single Deep Learning model does not require vast amounts of compute power; however, training whole populations of models until convergence quite often proves itself computationally unfeasible, requiring smart parallelization, zero-cost proxies and a whole repertoire of tools to try and lower the required compute. On the other hand, the performance of the different architectures will depend on all the facets of our task (amount and complexity of the data, inductive biases provided, data augmentation and/or adversarial training schemes…); thus, the performance of the optimizers employed to explore this hyperspace of architectures will be task-dependent as well. In particular, we are concerned with scarce but complex data regimes, like the characterization of arctifacts in CT images, and the interaction of NAS with Ensemble Learning techniques that aim to improve the generalization capabilities of the resulting models by combining the contributions of several individual learners within a population into a single, more robust prediction. Moreover, Ensemble Learning allows to limit the search to shallower models (vastly lowering computational expense) because it provides the necessary complexity to model the tasks by parallelizing the computations instead of serializing them. A novel NAS optimizer and a hybrid NAS-Ensemble Learning method were developed to test these dynamics, and the analysis carried out pointed towards some destructive interaction between NAS and Ensemble Learning driven by the validation overfitting produced by the data scarcity. These developments and subsequent findings were condensed into a congress contribution, submited to AutoML2024 with the title Transfer Learning by Ensemble Learning and Swarm Intelligence-based Neuroevolution, explained in further detail in the following short video:
Another important caveat of NAS is the increasing complexity as the space to explore broadens. Exploring simple, cell-based spaces like NAS-Bench 101 is relatively easy but completely unable to yield novel architectures, which are often required for very specific use cases. That is the motivation behind the einspace, a new architecture representation space based on context free grammar that allows the expression of any model that can be constructed with very simple operations (from transformers, to multi-layer perceptrons, to convolutional neural networks, to any hybrid in between) in a simple and clean manner. It was developed by the Bayesian and Neural Systems Group from the University of Edinburgh and it has the potential to yield high-performing architectures for very complex medical tasks. However, its novelty also means that many of the operations that are almost trivial in other search spaces are not yet defined for the einspace. In particular, we focused on crossover operators (allowing the generation of hybrid architectures and the reuse of functional blocks throughout the search) and the calculation of a distance metric between architectures (broadening the posibilities for performance analysis and driving the exploration-explotation tradeoff within the search). The combined efforts from the Hospital General Universitario Gregorio Marañón, the University Carlos III of Madrid, the University of Edinburgh and the Helmholtz-Zentrum Berlin für Materialien und Energie have yielded a novel method for calculating the shortest edit path between architectures with unprecedented speed, providing with an efficient tool to perform both distance analysis and crossover. This work is currently under review for the International Conference on Learning Representations (to be held April 23-27, 2026) under the title Evolutionary Architecture Search through Grammar-Based Sequence Aligment.
Resource Optimization
This activity evaluated and implemented different techniques that reduced memory consumption during the training phase of neural network models without precision loss. This made it possible to use higher-resolution images and avoided reducing image quality or using patches, as well as their associated problems of loss of detail or spatial context.
Results were presented at HeteroPar 2024 on the work «Strategies for memory management improvement in deep learning algorithms for contrast enhancement of high-resolution radiological images», Rodriguez, D.A., Sanderson, D., García-Blas, J., Desco, M. and Abella, M.
Several techniques available in the most popular frameworks such as PyTorch or Tensorflow have been evaluated. Some of them are: Gradient checkpointing, Automatic mixed precision, Data parallelism and Model parallelism. Evaluation was performed using both, a single GPU and multiple GPUs. Empirical testing proved that combining different techniques a memory reduction up to 20% using a single GPU and up to 70% using multiple GPUs can be achieved.
Using a single GPU
| Train Loss | Validation Loss | Memory (MB) | Time (s) | |
| Baseline | 4.0504 | 6.0083 | 1198.08 | 35.63 |
| Checkpointing (Chkp) | 4.0482 | 6.0592 | 942.08 | 35.15 |
| AMP | 4.8612 | 6.2515 | 583.68 | 34.02 |
| AMP + Chkp | 4.8612 | 6.2761 | 460.80 | 34.30 |
Using multiple GPUs
When using multiple GPUs there two different scenarios: Data parallelism, wherethe model is replicated among the different devices and the training data is splitted. And Model parallelism, where in contrast to Data parallelism, the model is divided and each part is sent to a different device, while the data is processed sequentially.
Data Parallelism
| Train Loss | Validation Loss | Memory (MB) | Time (s) | |
| Baseline | 4.0504 | 6.0083 | 1198.08 | 35.63 |
| Data Parallelism (DP) | 2.2267 | 6.1418 | 1290.24 | 12.97 |
| DP + AMP | 2.4507 | 6.2196 | 636.00 | 13.46 |
| DP + Checkpointing | 2.1602 | 5.8934 | 798.72 | 15.24 |
| DP + AMP + Chkp | 2.1586 | 6.2355 | 430.08 | 12.98 |
In Data parallelism, training loss is smaller as we are training each model with a subset from the original dataset. Therefore, we also reduce execution time.
Model Parallelism
| Train Loss | Validation Loss | Memory (MB) | Time (s) | |
| Baseline | 4.0504 | 6.0083 | 1198.08 | 35.63 |
| Model Parallelism (MP) | 4.5295 | 5.9778 | 732.16 | 53.29 |
| MP + AMP | 4.6431 | 5.7533 | 399.36 | 40.69 |
| MP + Checkpointing | 4.4820 | 5.6649 | 614.40 | 57.32 |
| MP + AMP + Chkp | 4.6431 | 5.7533 | 394.24 | 57,26 |
Unlike Data parallelism, execution time is increased due to communication overhead, however, memory consumption is highly reduced as the model is devided among the different devices.